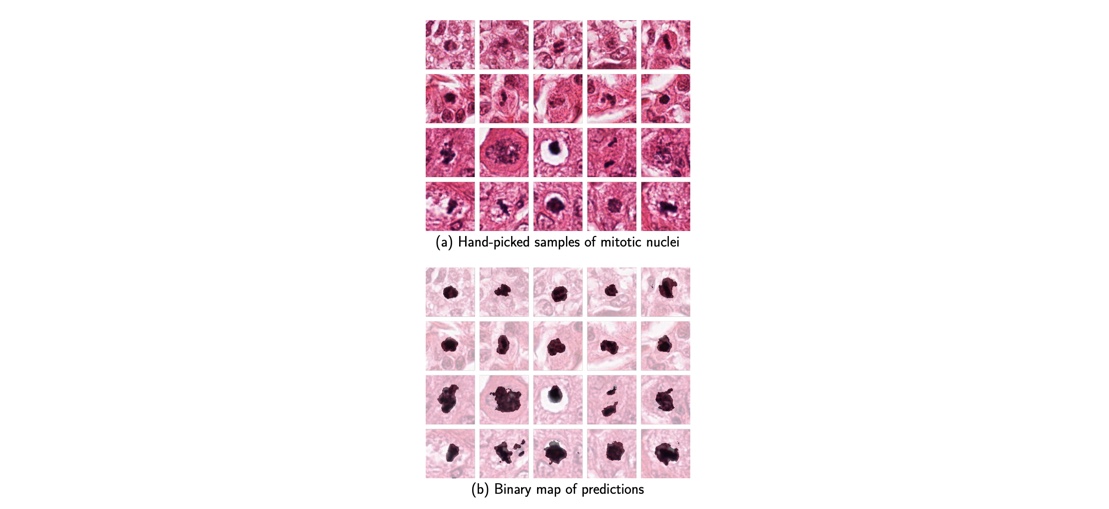
A Mitosis is a pretty rare phenomenon and and when its size is compared to the size of the breast cancer histology images, we are able to see why it is a needle in a haystack kind of a problem. Add to that the varied shapes it occurs in, it becomes even more challenging. We started with the Theano library and implemented a CNN model which was derived from the state of the art paper having the same title. Since, it did not work well because of the skewed data set, we tried to use Stain Segmentation [ Work of Abhishek Vahadne and team at IITG]. It had little to no effect on the accuracy of our results. We finally found out in January, 2016 that our network had the problem of dead weights due to random initialization. This was only possible because the CS231N lectures came out in December, 2015 and I used to follow them. We were very lucky that this problem was discussed in CVPR 2015 and therefore it was mentioned in the Stanford lectures. We also incorporated some other newer learning rules like AdaGrad and Adam update, which helped us omit tuning the learning rate. After this our model worked and we were able to achieve the results with a single CNN, where as the original paper used 2 cascaded CNNs. The F-score calculated for pixel classification was 0.51, with 0.42 precision and 0.63 Recall.
Repository and Poster Link: https://github.com/znck/mitosis-detection